Projects
Table of contents
Creating a new habitat
Creating new samples and DNA extracts
Importing sequence data
Creating a new habitat
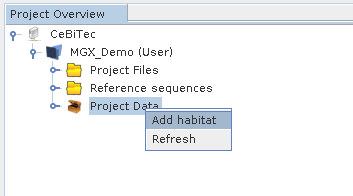
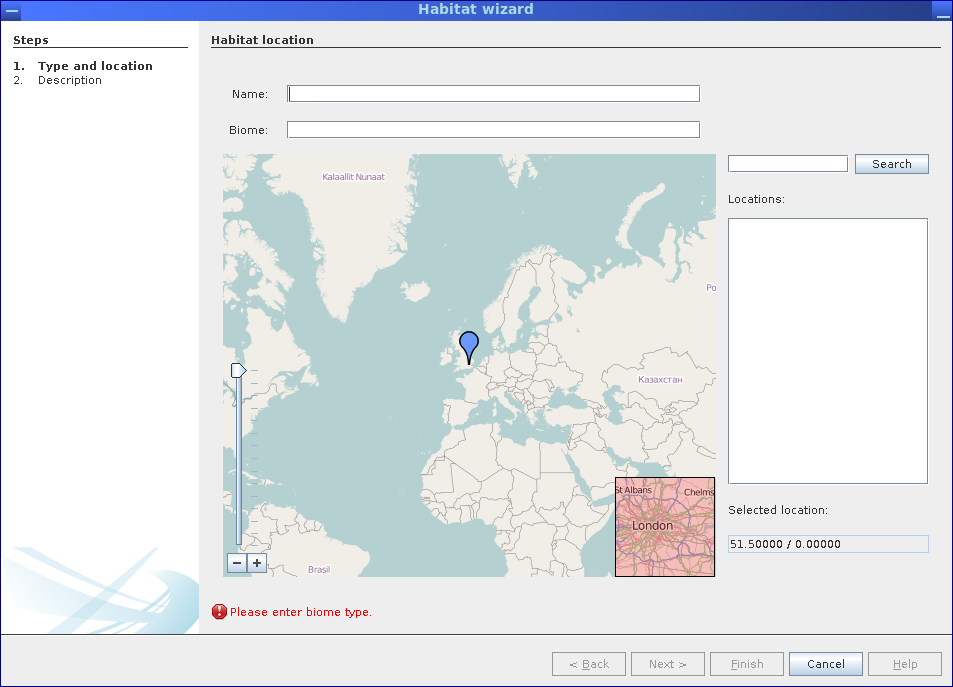
New habitats are defined choosing Add habitat from the context menu of the Project Data node. This will bring up a wizard allowing to select the corresponding geographical location as well as specifying a habitats name and biome type. In a second step, an optional description for this habitat may be entered, as well.
A new habitat can be added selecting the appropriate entry from the context menu of the Project Data node.
The habitat wizard allows to define new habitats specifying their location, name and biome type.
Creating new samples and DNA extracts
Samples and DNA extracts are created in the same way as habitats, except that samples are defined for habitats and DNA extracts are defined based on samples. Thus, the corresponding wizards are available from the context menu of the Habitat and Sample nodes, respectively.
Creating new samples
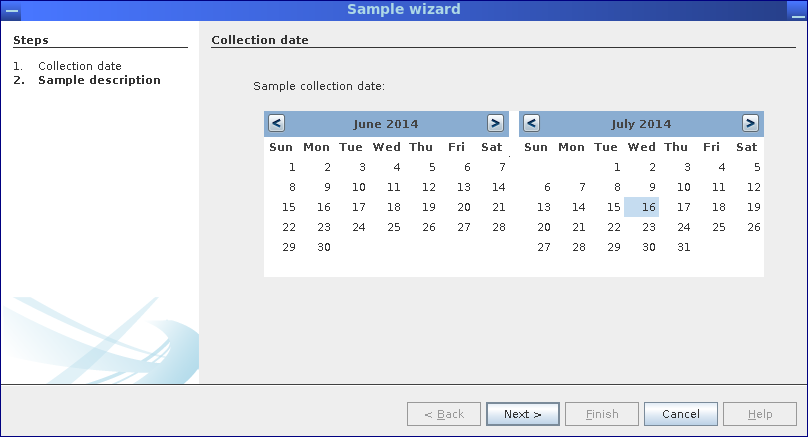
Step 1: Select sample date in sample wizard.
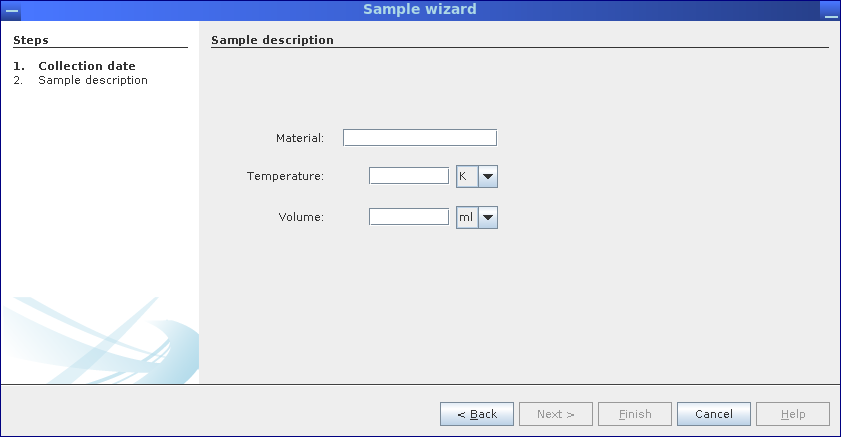
Step 2: Values for samples material, temperature and sampled volume/weight are required.
DNA extract
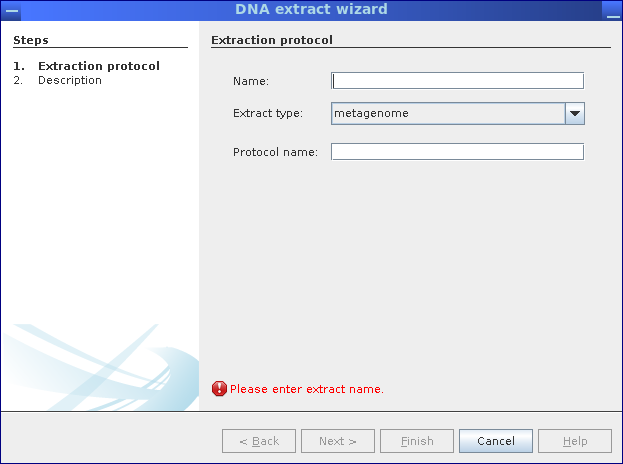
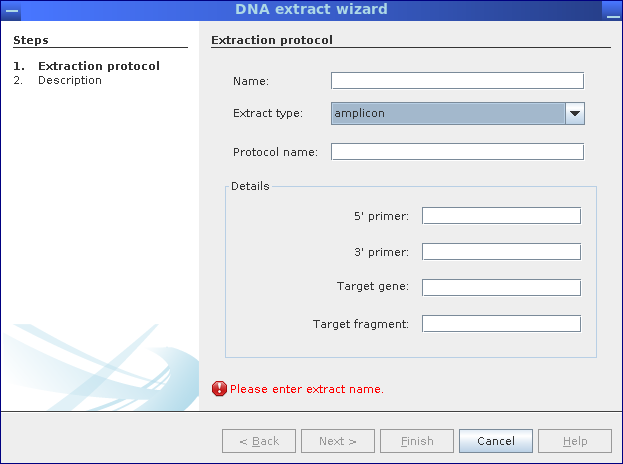
Step 1: Specify the type of DNA extract (metagenome, metatranscriptome, amplicon) and used protocols for DNA extractions.
Step 2: If needed, provide additional information.
- Amplicon: Primer names and corresponding target gene (fragment)
- Metatranscriptome: Select type of ribosomal RNA depletion methods
Importing sequence data
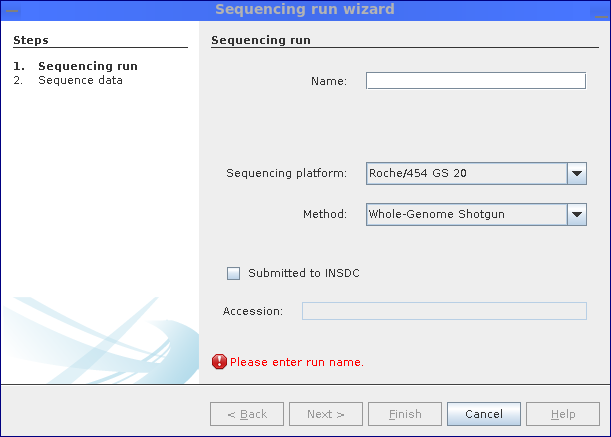
Finally, sequence data can be imported into the MGX project by creating new sequencing run objects; using the corresponding wizard, a name and details about the sequencing methodology such as platform and strategy can be deposited. Depending on whether single-end or paired-end/mate-pair data was selected, the wizard subsequently allows to select one or two files.
Give the large number of different sequencing adapters and barcoding schemes, MGX is unable to address all of them; hence, proper adapter removal, quality-based trimming (and filtering of e.g. host DNA should be performed before uploading datasets to MGX for analysis.
Step 1: Specify the employed sequencing platform and technology.
NOTE
Data which is already submitted to or obtained from public INSDC repositories (e.g. NCBI, EBI, DDBJ), the correspondingaccession numbercan be stored.
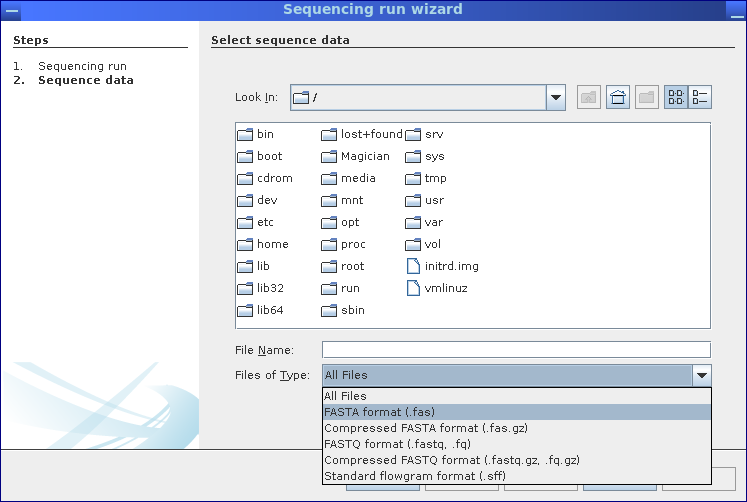
Step 2: Select file containing the sequence data.
MGX supports all commonly used file formats
- FASTA
- FASTQ
- SFF